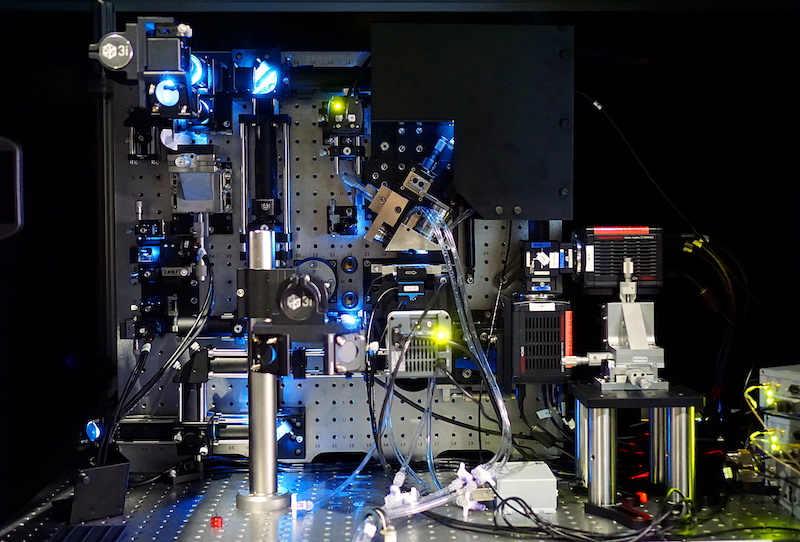
RCC is leading development of dedicated computing and storage infrastructure to handle the big data volumes expected to be generated by a revolutionary new microscope being installed at UQ’s Institute for Molecular Bioscience (IMB).
The Lattice Light Sheet Microscope (LLSM), a new modality for 4D imaging of live biological specimens ranging from individual molecules to small organisms, is expected to generate up to 7 TBs of imaging data per day.
This new type of microscope enables unprecedented imaging of live specimen without phototoxicity or photobleaching, enabling prolonged imaging of significant physiological or biophysical events. It was developed by 2014 Nobel laureate, Dr Eric Betzig of the Howard Hughes Medical Institute, Janelia Farm Research Campus, Virginia USA.
Installation of a LLSM at UQ is the result of successful funding bids to the Australian Research Council’s LIEF scheme and the Australian Cancer Research Fund by an international consortium of researchers led by IMB’s Professor Jenny Stow.
“The LLSM being installed at IMB is one of only a handful in the world and the third in Australia,” said Prof. Stow, “It will be used to generate exciting new insights into cancer and cell biology, immunology, virology, pathogens and bacterial infection, brain and organ development.”
RCC is leading development of dedicated computing and storage infrastructure, in collaboration with several institutes and schools at UQ, to support the LLSM and avoid critical bottlenecks in imaging as terabytes of data can be generated by a single microscope in a matter of hours.
Working closely with Prof. Stow's lab, RCC developed and is deploying visualisation software Phoebe specifically to help researchers deal with the large amount of data generated by the LLSM. Phoebe is a time-based, distributed, volumetric visualisation system that leverages UQ’s high performance computing, data storage and networking infrastructure. It is ideal for processing LLSM data as it provides researchers with the ability to visually survey very large LLSM data sets as if they were interactive movies. Researchers can easily move about anywhere in their data using Phoebe’s graphical front end from their desktop machines. This is accomplished by offloading the bulk of the data processing and storage onto UQ’s HPC infrastructure while sending the ‘visual’ data to the user.
Once captured on the LLSM, the data will be stored and made available via MeDiCI (Metropolitan Data Caching Infrastructure), a high performance data storage fabric RCC recently developed at UQ.
MeDiCI delivers seamless ultra-fast multi-site data access, saving researchers valuable time since manual movement of data is not required, and ensuring data integrity. Whether the researcher wishes to access their data from their desktop, via the OMERO image database (see below), or on one of the HPC options, such as Tinaroo or FlashLite, available at UQ, the data will appear without the need to move and duplicate multiple terabytes of data.
For management, storage, organisation, metadata, searching and availability, RCC in collaboration with UQ’s IMB, Queensland Brain Institute, and Centre of Microscopy and Microanalysis, has created an instance of the OMERO image database for use by all UQ researchers. OMERO allows secure image data storage and access via desktop clients or through web browsers. As well as sharing data within groups at UQ, OMERO allows easy and secure sharing with research collaborators, and public access where appropriate.
With supporting funding from the Australian National Data Service (ANDS), QCIF and RCC are working as part of a national consortium to develop digital tools to work with the LLSM. At UQ these include tools for visualisation and to interact with the massive image sets, as well as ensuring that the OMERO database can deal with and make easily available the microscope’s data. At Monash tools are being developed to create workflows and to pre-process LLSM data, as well as to capture important imaging metadata and created trusted data that researchers can analyse, share and re-use. Researchers at UNSW are enabling integration of these data and methods into the Characterisation Virtual Laboratory environment.
Together, these new initiatives in big data storage, advanced data processing and data access will ensure that UQ researchers can fully leverage the power of exciting new data sources from cutting-edge technologies such as LLSM, and facilitate the breakthroughs in biological and medical sciences they enable.



