The use of high performance computers FlashLite and Tinaroo cut usual compute times to a fraction of what it normally would have been for a study at the University of Queensland’s Institute for Molecular Bioscience on the genetic variation underlying human complex traits and disease.
For the study, the researchers analysed a large number of variants on the DNA sequence across many people. To perform the analysis, the team developed a Bayesian model that Dr Jian Zeng, a Postdoctoral Research Fellow at IMB, described as “a very challenging task computationally.”
“First, the data matrix we use often features tens of thousands of individuals with millions of variants, which requires hundreds of GB of memory to store,” said Dr Zeng.
“Second, the algorithm we used for the iterative sampling process often takes a long time to converge."
“More precisely, we needed to sample the effects of variants conditionally one-by-one using information from all individuals, and this sampling process proceeds over a number of iterations until convergence is reached. Thus, the computing time increases linearly with the number of variants and the number of individuals (sample size).”
To speed up the analysis, the research team used FlashLite for its large memory and parallel computing abilities in the sampling of each variant effect by using the HPC’s message passing interface (MPI).
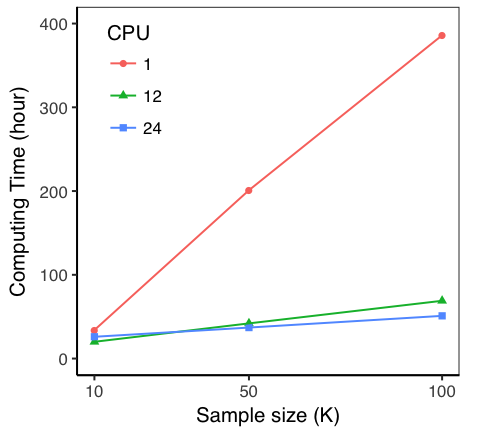
“We ran these analyses on FlashLite using 500 GB of memory and 24 CPUs for each human trait,” said Dr Zeng. “Although the MPI strategy allowed us to distribute the data across nodes, we knew that using a single node with large memory would be beneficial because our sampling process needs intensive communications among CPUs, which is slower across nodes. As shown in Figure. 1, the computing time reduced substantially when more CPUs were used.
“Using 24 CPUs, the analysis of each trait took up to a couple of days (actual time depending on the sample size). With multiple analyses running simultaneously, we managed to finish the analyses on dozens of traits in a week. Without FlashLite, it probably would’ve taken us months to run these analyses sequentially across nodes with normal memory capacity.”
RCC Director Prof. David Abramson said the IMB research project was the perfect match for FlashLite’s capabilities. “FlashLite has a large amount of memory per node, so it is possible to hold the whole data structure on a single node.”
The IMB researchers needed an even more powerful high performance computer for their next task: running genetic analyses on massive gene expression data.
“In this study, we needed to carry out a batch of analyses for tens of thousands of gene expression traits, although each analysis required only a small memory and one CPU. We used the HPC Tinaroo to run such massive analyses, where up to 1,000 jobs were run simultaneously,” said Dr Zeng.
“The analysis of each gene expression trait took a few hours, so all analyses were expected to be completed within a few days. We wouldn’t have been able to finish this study without the massive computing power of Tinaroo.”