GenomeSpace, a cloud-based framework to support genomics analysis through an easy-to-use Web interface, was added to Galaxy Queensland last month, thanks to the work of RCC.
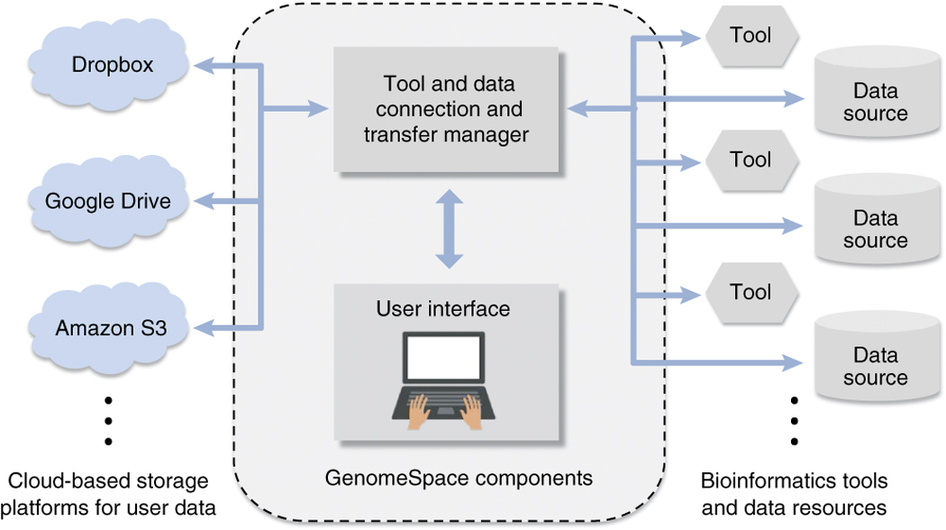
GenomeSpace enables easy integration and export of genomic data into and out of analysis pipelines. It is a community resource that currently supports the streamlined interaction of 20 bioinformatics tools and data resources.
Its key features include the ability to integrate with most life science software; compatibility with bioscience data formats; and the ability to act as a mediator for credentials, for private and public access to secure data.
RCC/QCIF/IMB eResearch Analyst Dr Nick Hamilton said a UQ post-doctoral researcher once told him he often spent literally half of his time solving data integration problems for genomic data sets before he could do the actual analyses. “GenomeSpace helps fix this problem, so it’s becoming increasingly popular and widely used across the biosciences.”
GenomeSpace’s inclusion on Galaxy Queensland, a public server for computational biomedical research, is part of Research Data Services’ (RDS) Life Sciences (Genomics) project. However, the initial concept and development of GenomeSpace started as a partnership between a handful of leading U.S. universities, including Harvard and Stanford (view the U.S. GenomeSpace website).
Nature Methods published a U.S./Israeli paper about GenomeSpace this year.
For more information about GenomeSpace, please visit its Australian website or contact RCC’s Dr Igor Makunin: i.makunin@uq.edu.au.



